FLP recombinase can be used to excise a transcriptional stop cassette from a Raeppli construct at a specific time during development.
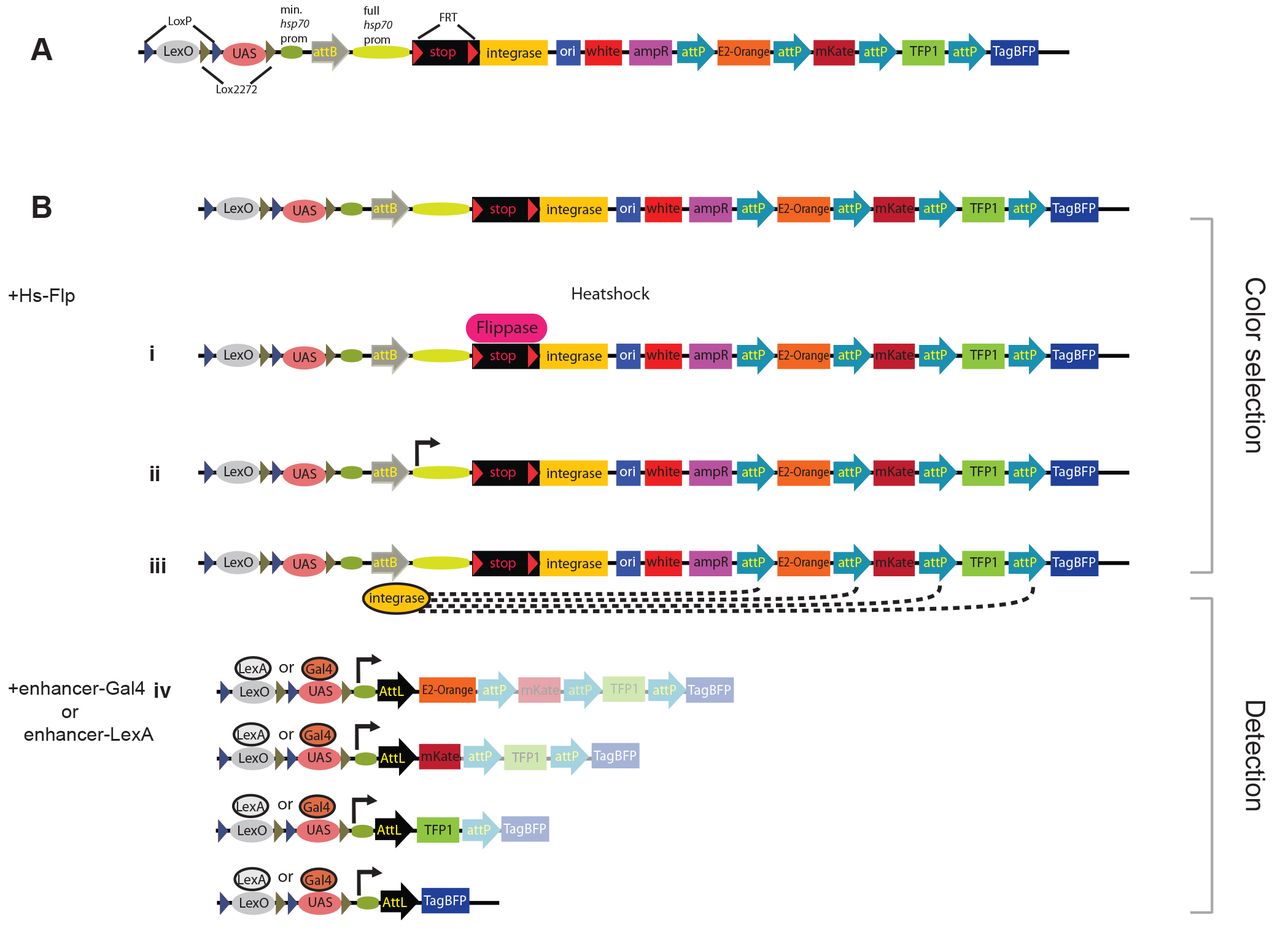
This allows the expression of phiC31 integrase, which catalyzes the further rearrangement of the construct to place one of four fluors under UAS or lexA control. These rearrangements establish mitotic clones, which can be visualized in specific tissues using lexA or GAL4 drivers to express the fluors.
Here is an example of the expression of the four fluors expressed in imaginal disc clones following rearrangement of a single Raeppli construct.
![]()
Using two copies of the Raeppli construct, one can track clones with ten different fluor combinations.
![]()
There are two Raeppli constructs: P{Raeppli-CAAX} expressing membrane-targeted fluors and P{Raeppli-NLS} expressing fluors with nuclear localization sequences.
Alternative loxP sites allow either the lexAop or UAS sequences to be removed optionally by exposure to Cre recombinase.
The method was described in Kanca et al. (2014), Raeppli: a whole-tissue labeling tool for live imaging of Drosophila development. Development 141: 472-480.

