We used two methods to determine the positions of the irradiation-induced breakpoints in the Dp(1;Y) chromosomes: Comparative Genome Hybridization (CGH) microarrays and PCR mapping relative to transposable element insertions.
CGH microarray mapping
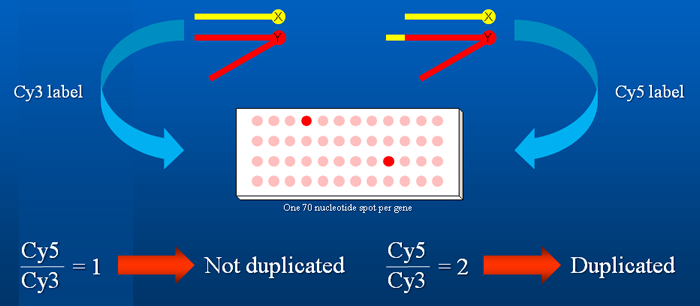
Dp(1;Y) chromosomes derived from In(1)BSC6 and In(1)BSC9 were characterized by CGH microarrays by Eric Spana at Duke University. In the microarrays, nearly every annotated gene was represented by a single ~70 bp oligonucleotide. In general, these microarrays provided two-gene breakpoint resolution: the breakpoint was positioned either within one of the adjacent genes or in the region between them.
PCR mapping
The breakpoints of the remaining Dp(1;Y) chromosomes were mapped relative to transposable element insertions spaced ~10 genes apart. When DNA samples from males carrying a Dp(1;Y) chromosome and an X chromosome with a transposable element insertion were PCR amplified using short extension times, primer pairs flanking the transposon insertion site produced a band only if the region of the insertion was present on the Dp(1;Y) chromosome.
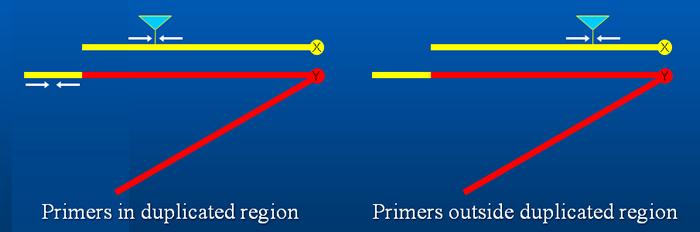
While we mapped the breakpoints with ~10 gene resolution, this technique can be used to narrow the positions of breakpoints even further. Resolution is limited by the density of transposon insertions in a particular chromosomal region.
Reference
R. Kimberley Cook, Megan E. Deal, Jennifer A. Deal, Russell D. Garton, C. Adam Brown, Megan E. Ward, Rachel S. Andrade, Eric P. Spana, Thomas C. Kaufman and Kevin R. Cook (2010) "A New Resource for Characterizing X-Linked Genes in Drosophila melanogaster: Systematic Coverage and Subdivision of the X Chromosome With Nested, Y-Linked Duplications" Genetics 186: 1095 - 1109.

